Study may open a new door for perio treatment
Published: 09/05/2018
A new UCLA-led study provides clear evidence that cellular messengers in saliva may be able to regulate the growth of oral bacteria responsible for diseases, such as periodontitis and meningitis.
The study's findings, which suggest that a body's cellular messengers play an important role in managing the amount of good and bad bacteria in the mouth, are published online and will appear in the May issue of the Journal for Dental Research.
Study authors Dr David Wong, professor of oral biology and associate dean for research at the UCLA School of Dentistry, and Dr Wenyuan Shi, chief executive officer and chief science officer at the Forsyth Institute, an oral health research institute in Cambridge, Massachusetts, asked the question of whether our RNA – the cellular messengers – can communicate with harmful bacteria in the mouth.
RNA acts as a messenger that transports DNA's instructions to other parts of the cell. Small regulatory noncoding RNAs, known as sRNAs, regulate our genes. A new class of sRNAs has also been discovered called tsRNA, which is transfer RNA-derived small RNA. tsRNA is found in human body fluids, including blood, tears and saliva.
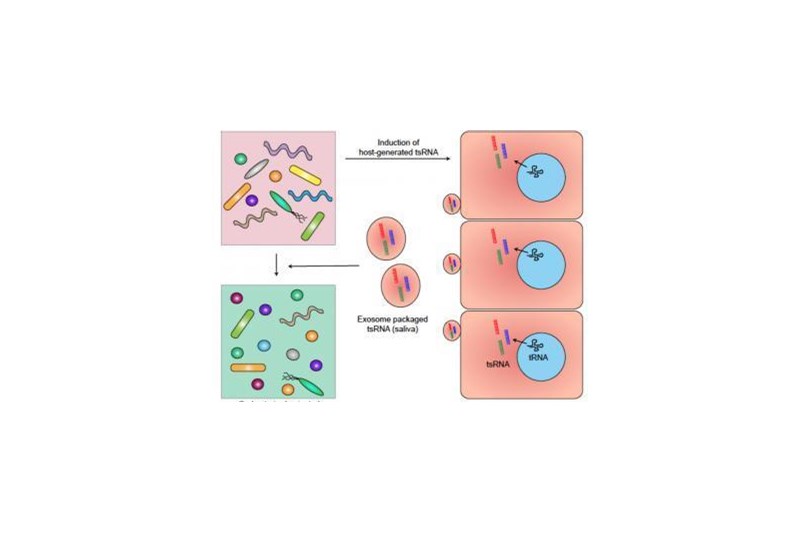
The research team began by analyzing salivary sRNAs and found many of them belong to tsRNA with their sequences matching the partial transfer RNA sequences of several Gram-negative oral bacteria -- bacteria that have a highly toxic outer layer that can cause periodontal disease. These salivary tsRNAs could potentially affect bacterial tRNA, a type of RNA molecule that helps decode a messenger RNA sequence into a protein and is required for bacterial growth. The Gram-negative bacterium used in the study to test this hypothesis was Fusobacterium nucleatum (F. nucleatum), the bacteria responsible for periodontitis.
The team showed that host cells respond to the presence of F. nucleatum by releasing specific tsRNA with sequence matching to tRNA of F. nucleatum. Furthermore, these released tsRNAs can inhibit the growth of F. nucleatum, but they have no effect on the growth of Gram-positive oral bacteria, such as Streptococcus mitis, a bacterium that responds to antibiotics.
'This study establishes that there is a clear channel of communication between RNA messengers and bacteria in our mouth,' said Wong, who holds the Felix and Mildred Yip Endowed Chair in Dentistry. 'Furthermore, we have shown that these messengers may play an important role in mediating interactions between bacteria and their host.'
Another significant study finding was the majority of tRNA bacteria sequences that show high sequence similarity with salivary tsRNAs came from antibiotic-resistant Gram-negative bacteria. This observation could lead to a better understanding of the mechanisms behind the growth of oral bacteria, resistance to antibiotics, and in-turn oral diseases, Wong said.
'Our findings could lead to new therapies to treat diseases caused by harmful bacteria,' said Shi, a former professor of oral biology at the UCLA School of Dentistry. 'As a convener for oral health and biotech experts, Forsyth is thrilled about collaborating with UCLA on this exciting research.'
For example, one of the hallmarks of periodontitis is a shift from mostly Gram-positive bacteria to mostly Gram-negative bacteria. With a better understanding of how Gram-negative bacteria grow, perhaps there is potential to reverse the growth or even kill Gram-negative bacteria.
'Our finding could potentially open a new door for prevention and treatment of periodontal diseases,' said Dr Xuesong He, a researcher at the Forsyth Institute and the co-first author on this study.
Author: Julie Bissett










.jpg?width=150&height=100&scale=canvas)